Sometimes I want to see the differences between a file (i.e. the thing on disk) and the Emacs buffer that “visits” the file. For example, before saving a file, I might want to check what changes will be saved to disk. Another thing that happens to me regularly is that a file gets changed on […]
Continue reading
For roughly the first ten years of my Emacs life, I simply opened Emacs each time I wanted to edit a file. In general, this worked fine. Of course it meant that I had to keep copies of my ~/.emacs file up to date on various machines, but as I wasn’t in the habit of […]
Continue reading
Most “ordinary” editors I know keep a list of the most recently opened files in their File menu. Emacs (of course) can have that too, through the built-in recentf package. Loading that package adds a submenu called Open Recent to the File menu. Alternatively, one can call M-x recentf or M-x recentf-open to open one […]
Continue reading
Unfortunately, just like most other software, Emacs can crash sometimes. Or maybe your whole system hung and you had to reboot. If such cases, it is good to know that Emacs makes backups of the files that you opened. To try and recover from the last backup you can use either of the following functions: […]
Continue reading
Something that may come up when sharing your Emacs screen in a video call: how to increase or decrease the font size? This turns out to be quite easy with the mouse scroll wheel: C-scroll will resize the font of the current buffer C-M-scroll will resize the font for all buffers. If you don’t like […]
Continue reading
A short one this time. For some questions — for example, if there are still unsaved buffers when you exit Emacs — Emacs will ask you to confirm by typing yes or no. While this helps against mistakes, I also find it annoying to have to type the whole word — even though we’re only […]
Continue reading
Back in 2010, as I started my postdoc at the Erasmus Medical Center in Rotterdam, I got introduced to R. Given that I was used to writing all my code in Emacs, it was obvious that I would write my R code there. I remember briefly looking into Tinn-R for the courses we would teach, […]
Continue reading
A few weeks ago, Bozhidar Batsov, writer of the Emacs Redux blog wrote a post on isearch, the function you use to search text in the current buffer, and how there is a lot more to it than just typing C-s to search for some text. I use C-s (isearch-forward) and C-r (isearch-backward) a lot. […]
Continue reading
In the previous three Emacs tips, I discussed various ways to select a window: other-window bound to the painful C-x o, which cycles from one window to the next; setting the mouse-autoselect-window variable to t for “focus follows mouse”; the windmove package that allows you to use a key prefix (super ctrl in my config) […]
Continue reading
Two tips ago, I mentioned the built-in windmove package as one way to make moving between Emacs windows easier. Both the awkwardness of the C-x o shortcut for the other-window command and the fact that it isn’t always clear which of the visible windows actually is the “other window”, call for a more intuitive way […]
Continue reading
Back in tip #4 about mouse-yank-at-point, I made a side note about the fact that I’m a long-time user of “focus follows mouse”: whenever I move the mouse cursor over a window, that window gets the focus. Without my having to click in that window. In the previous tip, I explained basics of Emacs’s window […]
Continue reading
Let’s start with a reminder of Emacs terminology. In Emacs parlance, a “frame” is what we normally call a window: the thing you can move around with your mouse (or with keys if you love tiling window managers). A “window” in Emacs, is part of an Emacs frame. By default, a frame starts with a […]
Continue reading
Working in Emacs often means you end up with loads of buffers: one for every file you opened, but also others for e.g. Magit, Tramp connections or flycheck. This is fine, in principle. Emacs doesn’t really care. However, as the number of buffers grows, you may find that switching between buffers becomes difficult as you […]
Continue reading
The rainbow-delimiters package contains a minor mode that highlights various sorts of brackets, braces, parentheses, etc. according to the depth in which they are nested. Obviously, this can be very helpful when editing lisp code (or your .emacs file), but I have also found it to be helpful in other languages, as shown in the […]
Continue reading
One of the features that Emacs has to help you with your code is show-paren-mode. By default this global minor-mode is turned on, which means that Emacs will highlight matching parentheses. Take the following R expression, for example: myfun <- function(parA, parB) { paste(“Doing nothing”) } Here, if the cursor is between the last n […]
Continue reading
This week, I’ll discuss how to easily comment parts of your code.
Continue reading
A few days ago, on the 23rd of February, the Emacs maintainers released the latest major version of Emacs: v30.1 (new Emacs major versions always seem to wait for the .1 minor version before being released publicly). This release has a couple of interesting new features and updated behaviours. As usual, Mickey Petersen has prepared […]
Continue reading
By default, Emacs doesn’t show line numbers in the left margin of the screen, like many other IDEs do. To me this feels especially clumsy when I’m working on a script or a LaTeX document and running the script or compiling the document points out an error on a certain line. Luckily, the built-in display-line-numbers-mode […]
Continue reading
By default, the point (the cursor in Emacs-speak) is shown as a flashing block. And for years that was what I used. The flashing was enough for me to locate the cursor. Back in July of last year, however, I started to look for something that showed the cursor location more clearly. I don’t exactly […]
Continue reading
Today’s tip is about managing superfluous spaces (in the sense of whitespace). For that, I use the following two functions: The function delete-horizontal-space (bound to M-\) removes all whitespace (spaces and tabs) around the point (Emacs-speak for “the cursor”). The function cycle-spacing (bound to M-SPC) is a “smarter” version of the above. It manipulates whitespace […]
Continue reading
This week, another “tip” that is both a tip and some background information. Emacs has been called the self-documenting editor. The first time may very well have been in the 1981 paper by Richard Stallman titled EMACS the extensible, customizable self-documenting display editor (https://doi.org/10.1145/872730.806466). The Emacs manual describes it in the following way: Self-documenting means […]
Continue reading
This week’s “tip” is not so much a tip as it is a bit of background on how Emacs works. A bit of extra knowledge that will hopefully help you in your Emacs journey. Although you may not realise it, many of your interactions with Emacs consist of calling functions. Keybindings and menu items call […]
Continue reading
After enabling diff-hl-mode by Github user dgutov any lines with uncommitted changes in a (version controlled) file will be highlighted in the margin. In large files, for example, this makes it easy to spot the sections that you have edited. Here is how I enable diff-hl-mode for all files: ;;;;;;;;;;;;;;;;;;;; ;; Enable diff-hl-mode (https://github.com/dgutov/diff-hl) to […]
Continue reading
Next week, I will be back at the Erasmus Medical Centre in Rotterdam to teach the EL016 Linux for Scientists course, which is part of the NIHES MSc programme in genetic epidemiology. Running for two full days, the course aims to teach students with a background in the life sciences to work on a (remote) […]
Continue reading
Several lint tools and Git pre-commit hooks require that files end with a newline character. This is easy to achieve in Emacs through the require-final-newline variable. By default this variable is nil, but when set to t Emacs will automatically add a final newline (if it isn’t already there) when saving a file. I have […]
Continue reading
The past few weeks have been terribly busy with all kind of end-of-year things, so I didn’t manage to find time for posting new Emacs tips. Sorry about that 😇. So, without further ado, here’s a new one. This week’s tip is about pasting with the mouse. By default, Emacs acts like most other applications: […]
Continue reading
This week’s tip is about balancing delimiters like parentheses, brackets, quotes, etc. The built-in electric-pair-mode does the following then enabled (copied from the manual, emphasis mine): Electric Pair mode, a global minor mode, provides a way to easily insert matching delimiters: parentheses, braces, brackets, etc. Whenever you insert an opening delimiter, the matching closing delimiter […]
Continue reading
Tramp is a built-in Emacs “package” that allows you to edit files on remote servers with your local Emacs instance. But it doesn’t only do that, it can also be used to edit files as root via sudo. To achieve this use C-x C-f as you normally would to open a file. However, instead of […]
Continue reading
My Emacs journey and the first weekly Emacs tip
Continue reading

In a little more than two weeks from now I will be at the ErasmusMC university medical centre to teach the “Linux for Scientists” course. The EL016 course, as it is officially called, consists of two full days of teaching and practice and is scheduled on January 31st and February 1st, 2023. It is part […]
Continue reading
Posted on 13 January 2023 by Lennart Karssen

Time flies! Since the end of July I have been working not from the Netherlands as usual, but from Uganda. Now, this “expat period” is coming to an end and my family and I will be back in Den Bosch in January. All in all, I think that my working remotely has not affected the […]
Continue reading

In two weeks’ time, on Wednesday 26th and Thursday 27th of January, I will be two full days at the Erasmus Medical Centre, Rotterdam. I will be teaching the course Linux for Scientists for students of the Netherlands Institute for Health Sciences (NIHES) MSc programmes.
Continue reading
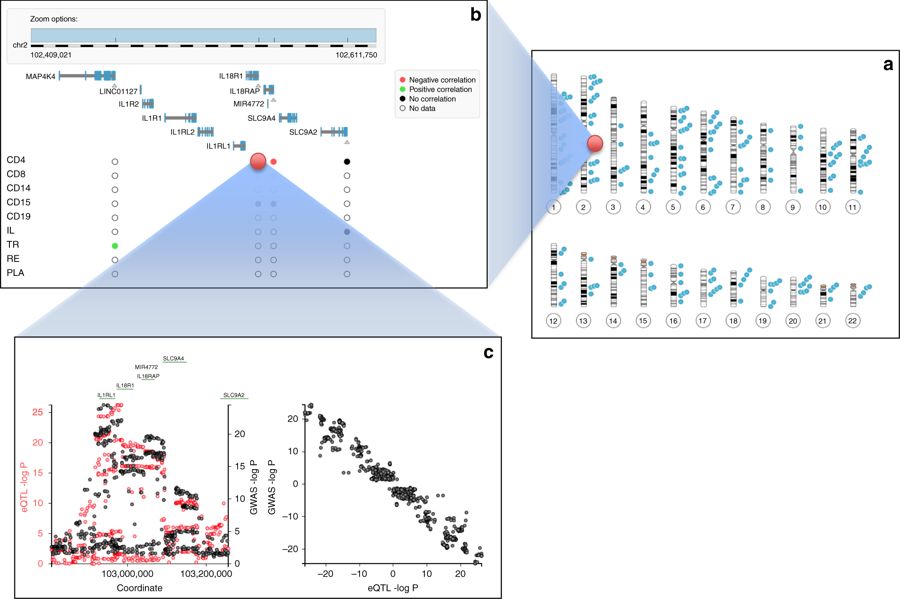
This database stores tens of billions of associations of genetic variants with human traits, investigated by the scientific community in hundreds of studies. Knowledge of such associations allows for better understanding of human genetics and biology and can contribute to the diagnosis, prevention and treatment of diseases. The results of the work were published in […]
Continue reading
Immunoglobulin G (IgG) that plays central role in adaptive immunity is the most abundant immunoglobulin found in human plasma. All IgG molecules are N-glycosylated. N-glycosylation of IgG contributes to the maintenance of its structure, stability, and solubility. In a large collaborative study, we analysed association between genomes and glycosylation of immunoglobulin G (IgG) in more than […]
Continue reading
A team of six, with two authors affiliated with PolyOmica, has received a 2020 prize in Clinical Science from the International Society for the Study of the Lumbar Spine (ISSLS) for a study that found a significant causal effect of BMI on both back pain and chronic back pain1. Back pain is the #1 cause […]
Continue reading
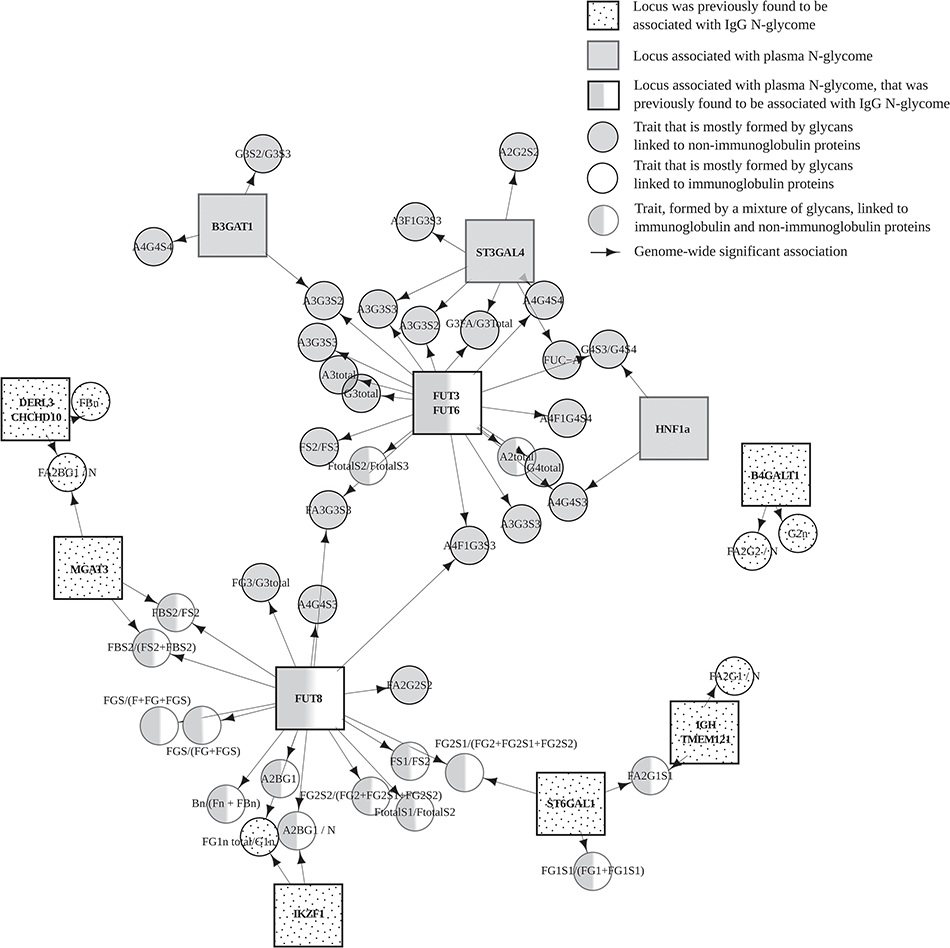
Glycosylation – addition of carbohydrates – is a common cotranslational and posttranslational modification of proteins. Attachment of different glycans to the same protein can give rise to hundreds of its glycoforms. Different glycosylation of the same glycoprotein changes its physical properties as well as its biological function. In recent years, changes in different glycan abundances were associated with various diseases, including many of these […]
Continue reading
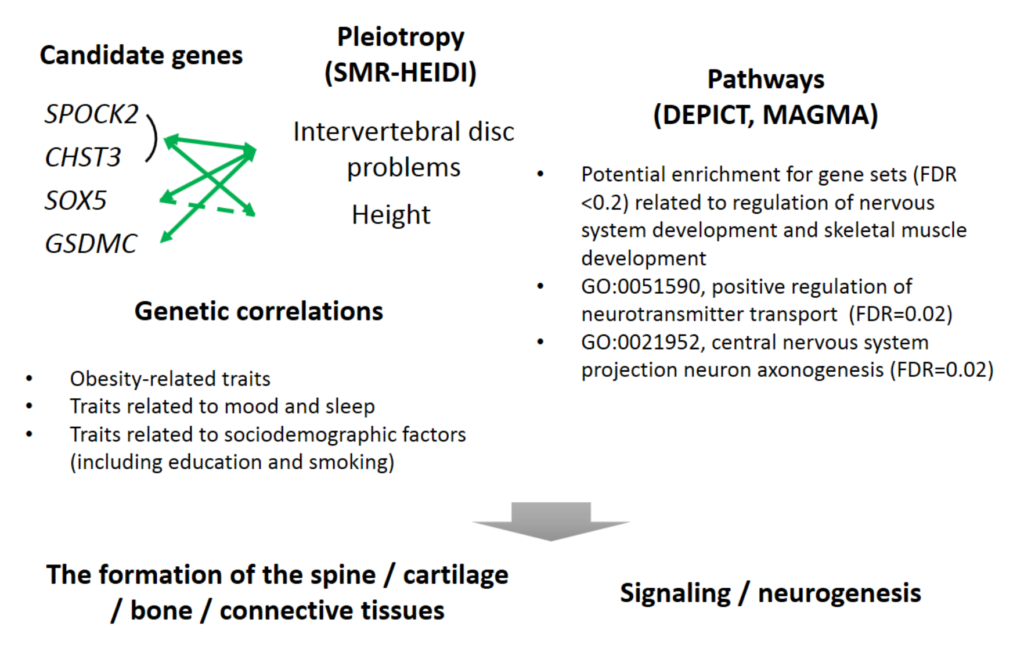
Back pain is the number one cause of years lived with disability worldwide and one of the most common reasons for health care visits in developed countries, yet surprisingly little is known regarding the biology underlying this symptom. In our previous study1, published in Sept 2018, we identified three novel genetic variants associated with chronic […]
Continue reading
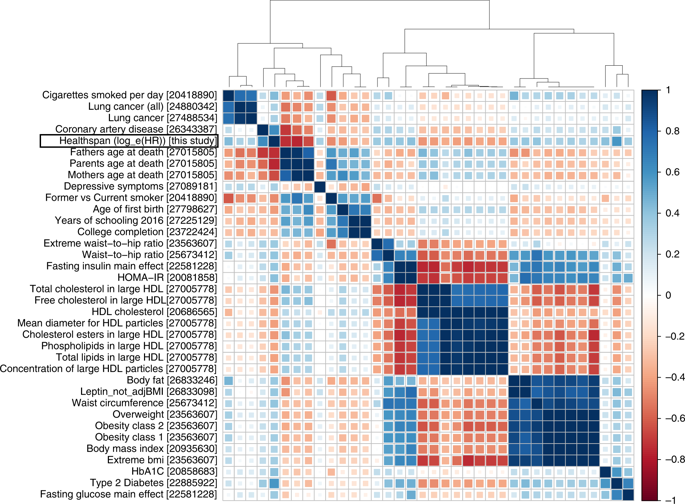
Aging populations face diminishing quality of life due to increased disease and morbidity. These challenges call for longevity research to focus on understanding the pathways controlling healthspan. A new study, led by dr. Peter Fedichev (Gero LLC) and prof. Yurii Aulchenko (PolyOmica), used the data from the UK Biobank to understand biology of healthspan1. We observed that the […]
Continue reading
Posted on 12 December 2018 by Lennart Karssen
The Clarivate Analytics list of Highly Cited Researchers for 2018 identifies scientists who have demonstrated significant influence through the publication of multiple highly cited papers in the last decade. PolyOmica’s Chief Scientist, Yurii Aulchenko , was selected for his substantial influence across several fields in the period 2006–2016. Approximately 6,000 researchers are named Highly Cited Researchers in […]
Continue reading
Adding covariates can either increase or decrease the power of finding an effect of a predictor on the outcome of interest. In a recent publication in GigaScience we used knowledge of biochemical networks to select covariates for conditional genetic association analysis. We demonstrated that the power depends on whether genes and environment play along or […]
Continue reading
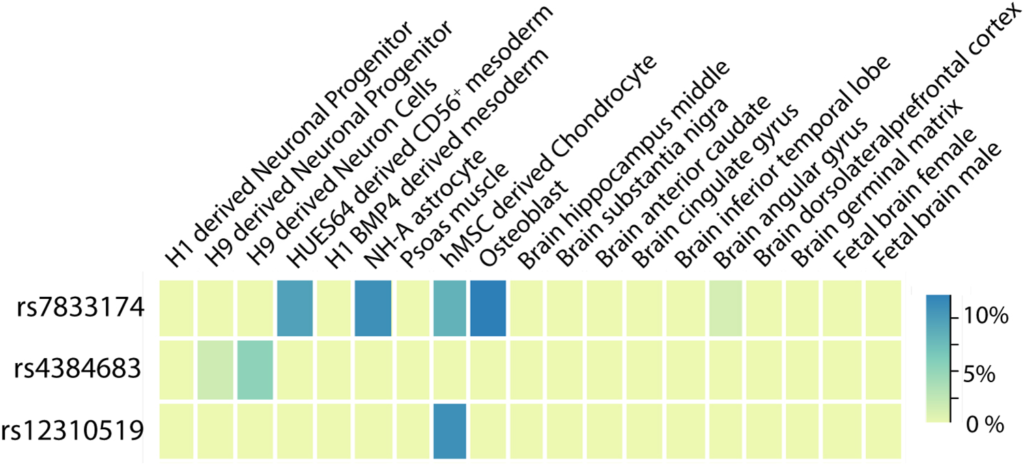
Back pain is the #1 cause of years lived with disability worldwide and one of the most common reasons for health care visits in developed countries, yet surprisingly little is known regarding the biology underlying this symptom. A new study, led by Pradeep Suri of the Department of Veterans Affairs in Seattle, Washington (U.S.A.) and […]
Continue reading
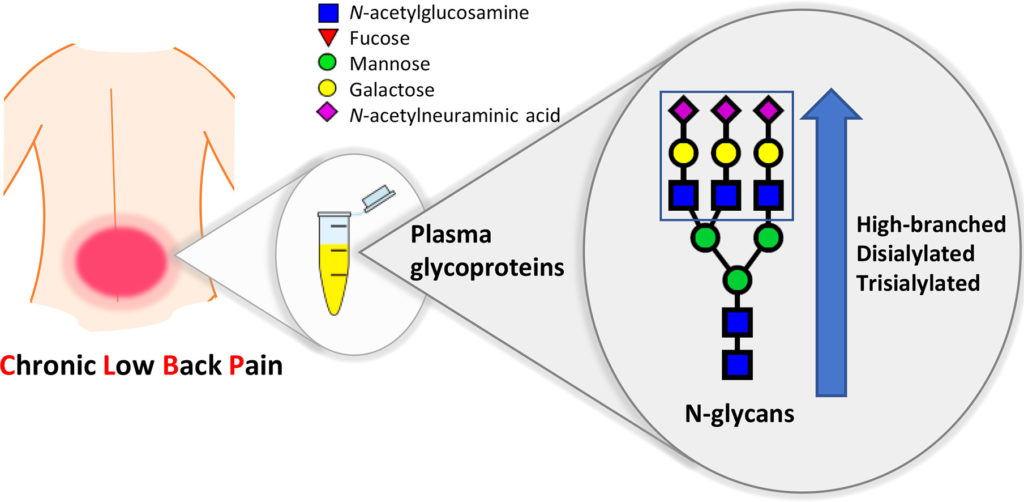
Last Thursday, July 5, 2018, a manuscript1 reporting results from a large multicentric case-control study of patients with chronic lower back pain (CLBP) was published in Biochimica et Biophysica Acta (BBA) – General Subjects. The work was funded by the EU FP7 project PainOmics in which we participate. The study of plasma N-glycome composition in patients […]
Continue reading
Inflammatory bowel disease (IBD) is a group of inflammatory conditions of the colon and small intestine. Crohn’s disease (CD) and ulcerative colitis (UC) are the principal types of inflammatory bowel disease. IBD was virtually unheard of in the early nineteen hundreds. Since then it’s incidence (as well as that of a series of other inflammatory diseases including asthma) has dramatically […]
Continue reading

From March 26–28, 2018 the IMforFUTURE International Training Network (ITN), in which PolyOmica is a partner, will have its first network meeting. The meeting will be held at the BioCentar in Zagreb, Croatia, and aside from various committee meetings and social events, the meeting will consist of training sessions for the fellows enrolled in the […]
Continue reading

On Monday March 5 2018, I will be teaching at the NIHES course “An Introduction to the Analysis of Next-Generation Sequencing data” at the Erasmus Medical Centre in Rotterdam. In this one-week course students will learn the basics of how NGS data is generated and how to analyse this type of data. PolyOmica’s contribution to […]
Continue reading

In PolyOmica, we some times need to train people in the basics of genetics and genomics. While training people in basics is fun, it is quite time consuming, and is not exactly our company’s core activity. For a long time, we thought that it would be great to have a massive open online course (MOOC) […]
Continue reading

Back in 2014 we initiated the Research Summer School in Statistical Omics with the help of many friends and colleagues, and with the support (financial and other) from various sources, including the EU-funded MIMOmics project, in which we participate, and the EU-funded Integra-Life project. In a period of two weeks we immersed 13 students (and […]
Continue reading

From 20 till 27 August, the EU FP7 consortium MIMOmics in which PolyOmica is a partner, organises a Summer School including a Workshop in Omics Studies. The goal of the School is to train a new generation of statistical omics scientists with a biological and biotechnological background to enable them to understand and to apply […]
Continue reading
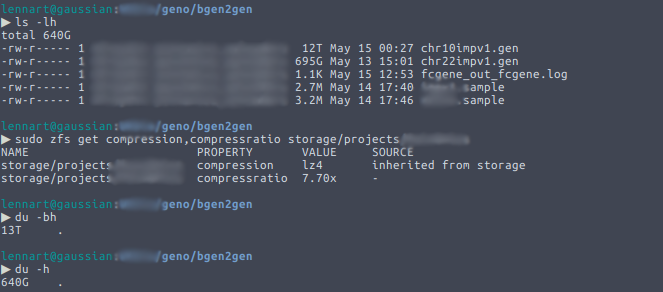
One of the great things of ZFS is its built-in compression capability. Enabling compression on a ZFS filesystem allows you to save a significant amount of space, especially in a field like ours where one sometimes needs to use large plain text files. It is completely transparent: the user doesn’t notice anything. Well… unless they […]
Continue reading

Next week (starting Monday January 30 2017) Yurii and I will be in Rotterdam at the Erasmus University Medical Center where we will teach in the “Advances in Genome-Wide Associations” course of NIHES. This course is part of the Master’s programme in Health Sciences and is compulsory for those who follow the specialisation “Genetic Epidemiology”. […]
Continue reading












